Simulation of pandemic spread
Title: Simulation of pandemic spread
Author: Daniel Kopecký
Method: Agent-based model
Tool: NetLogo
Contents
Introduction and problem definition
Not so long ago we had the COVID-19 pandemic, which showed us the shortcomings in dealing with this type of problem. Pandemic propagation simulation can be a key tool to model, analyse and predict the evolution of a pandemic. This model deals specifically with viral diseases. The aim of this simulation is to be able to predict the spread of a pandemic virus, whereby using appropriate values, it can simulate the approximate development of a pandemic in the Czechia.
Method
An agent-based model in NetLogo is used to simulate the spread of the pandemic. This allows us to code our own scenario using different variables and helps us to get closer to the real pandemic evolution.
Model
The model contains a map of the Czechia, where the selected population is randomly scattered at the beginning of the simulation. The inhabitants can move freely within the entire rendered territory. The selected population is already infected and by running the simulation they can infect other uninfected citizens. The model offers to create a custom scenario where the user sets their own variable values. It is also possible to choose from preset viruses that set the variables for the user.
Environment
The model is reserved for the Czechia. This was achieved using a silhouette of the Czech Republic that was uploaded to NetLogo using this code: import-pcolors "cesko.png".
Agents
People are represented by agents who have three different colors. People who are healthy, but not immune, are green. These people can also be infected by infected individuals. The red colour is used for people who are infected. So these people can spread the virus, get better or die from the virus. Individuals who have had the virus and are now immune to the virus are blue.
Movement
People move randomly around the territory of the Czechia. During their movement they may meet other individuals from whom they could potentially be infected.
Spread of infection
The original infected individuals can infect any individual that comes into their vicinity. The variables qurantine.effort, trasmission.rate, and the number of infected around an uninfected individual affect whether an individual becomes infected. If an individual has already been infected but has recovered, this means that it has immunity and therefore cannot be re-infected. Immunity can be turned off when setting up the model.
Recovery from infection
Individuals who are infected have a chance to recover, which is set by the recovery.rate variable. If they recover and immunity is on, the individual's color will change to blue. If immunity is off, the color of the individual will change to green.
Death
If an individual is infected, there is a chance that they will die. This is affected by two variables, namely healthcare.capacity and infected-mortality. Healthcare.capacity is the spare capacity of healthcare facilities. The value ranges from 0 to 1 and represents the maximum % of the population that can be hospitalized at one time. If the % of infected in the population exceeds the capacity limit of the healthcare facilities, the probability of an individual dying rises.
End of the simulation
The simulation ends when the virus stops spreading and the number of infected drops to zero, or when the number of infected equals the population size, or when the virus kills off the entire population.
Predefined viruses
The model offers the possibility to choose a predefined virus or to choose a custom scenario where the user defines their own variable values. Once a virus is selected, its transmission rate and mortality rate are set.
Covid-19
Covid-19, caused by the novel coronavirus SARS-CoV-2, emerged in late 2019 and quickly developed into a global pandemic. It primarily spreads through respiratory droplets, causing a range of symptoms from mild respiratory issues to severe pneumonia, with a heightened risk for older adults and those with underlying health conditions.
R0 = 0.71 [1]
Fatality rate = 0.0004 [2]
Recovery rate = 9 [3]
Immunity = True[4]
Spanish Influenza
The Spanish Influenza, which occurred in 1918, was an exceptionally deadly H1N1 influenza A virus that caused a devastating pandemic.
R0 = 1.2 - 3.0 [5]
Fatality rate = 0.03 [6]
Recovery rate = 7-30 (cannot be exactly found) [7]
Immunity = CANNOT BE FOUND
Seasonal Influenza
Seasonal Influenza, or the flu, is an annual respiratory illness caused by influenza viruses. It typically circulates during the colder months and can lead to fever, cough, and muscle aches.
RO = 0.9 - 2.1 [8]
Fatality rate = 0.000005[9]
Recovery rate = 7[10]
Immunity = False (own experience :-) )
Measles
Measles is a highly contagious viral infection, primarily affecting children, characterized by a distinctive rash and fever.
R0 = 12-18[11]
Fatality rate = 0.001[12]
Recovery rate = 7-18 [13]
Immunity = True [14]
SARS
SARS (Severe Acute Respiratory Syndrome) emerged in 2002, caused by a coronavirus (SARS-CoV), resulting in severe respiratory distress.
R0 = 2.7[15]
Fatality rate = 0.096[16]
Recovery rate = 10-14 [17]
Immunity = True [18]
Ebola
Ebola, a severe and often fatal viral hemorrhagic fever, gained global attention during outbreaks in Africa. It causes internal bleeding and multiple organ failure, with a high mortality rate.
R0 = 1.95[19]
Fatality rate = 0.5[20]
Recovery rate = 10-14 [21]
Immunity = True [22]
Variables
- init-population - Population at the beginning of the simulation
- init-infected - Number of infected at the start of the simulation
- recovery.rate - Rate of recovery of infected individuals
- init-immune - Number of immune individuals at the beginning of the simulation
- quarantine.effort - Quarantine effort (affects the chance of infecting an individual)
- transmission.rate - Rate of virus transmission between individuals
- infected-mortality - Virus mortality rate
- healthcare.capacity - Capacity of health facilities (affects the rate of death of individuals)
- immunity? - Turns immunity on or off
UI
The user interface consists of setup and go buttons. These are used to prepare all necessary data and values before starting the simulation and to start or pause the simulation itself.
A switch that enables/disables immunity, which affects whether individuals in the simulation can be immune to the virus after they have recovered from it.
Also from various sliders that set the values of variables that are then used within the simulation run.
The population slider sets the initial number of individuals that appear in the simulation. We can choose from values 1-1000.
Slider to set the initial infected individuals.
A slider to set the rate at which individuals heal.
Slider to set the initial state of immune individuals.
Quarantine effort, which tells us how individuals are trying to comply with the quarantine.
The transmission rate of the virus between individuals.
The mortality rate of the virus when an individual is infected with it.
And the capacity of health facilities, which is reflected in the rate of spread of the virus if it is exceeded.
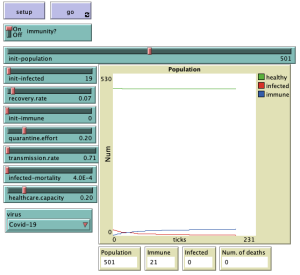
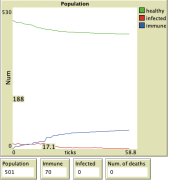
Results
Here we can see on the graph the results for the individual diseases that are selected, with the quarantine effort set to 0.2 and the capacity of the health facilities also set to 0.2.
For all simulation results shown above, the default settings for the selected virus were used. The population was set to 501, the number of infected individuals to 19, and the number of individuals with immunity to 0.
Conclusion
The model in this simplified version can simulate the development of various pandemics within the Czech Republic. Its main advantage is the limitation of the territory that follows the borders of the Czech Republic and also the preset values for different viruses, which are based on real data. We must also not forget the customization option, thanks to which the user can create tailor-made virus simulations. Creating the model itself was not as challenging as searching for all the background information for each virus. As part of the extension, it would be possible to add additional predefined viruses or to add additional refinement parameters that would affect the course of the entire simulation for a given virus.
NetLogo File
Below I attach the source file together with the map of the Czech Republic, which must be in the same directory as the NetLogo source file.
File:Virus sim.nlogo
![]()
Sources
- ↑ COVDATA. [online]. [citováno: 11.1.2024]. Dostupné z: https://www.covdata.cz/cesko.php
- ↑ Edouard Mathieu, Hannah Ritchie, Lucas Rodés-Guirao, Cameron Appel, Charlie Giattino, Joe Hasell, Bobbie Macdonald, Saloni Dattani, Diana Beltekian, Esteban Ortiz-Ospina and Max Roser (2020) - "Coronavirus Pandemic (COVID-19)". Published online at OurWorldInData.org. Retrieved from: 'https://ourworldindata.org/coronavirus' [Online Resource]
- ↑ HABTAMU TAMIRU, Desiyalew, Abebaw GEDEF AZENE, Gebeyaw WUDIE TSEGAYE, Kebadnew MULATU MIHRETIE, Samuel HUNEGNAW ASMARE, Wudneh AREGA GETE a Simachew ANIMEN BANTE, 2023. Time to recovery from covid-19 and its predictors in patients hospitalized at Tibebe Ghion Specialized Hospital Care and Treatment Center, a retrospective follow-up study, North West Ethiopia. Global Health [online]. roč. 2023, s. 1–10. Získáno z: doi:10.1155/2023/5586353
- ↑ STEIN, Caroline, Hasan NASSERELDINE, Reed J SORENSEN, Joanne O AMLAG, Catherine BISIGNANO, Sam BYRNE, Emma CASTRO, Kaleb COBERLY, James K COLLINS, Jeremy DALOS, Farah DAOUD, Amanda DEEN, Emmanuela GAKIDOU, John R GILES, Erin N HULLAND, Bethany M HUNTLEY, Kasey E KINZEL, Rafael LOZANO, Ali H MOKDAD, Tom PHAM, David M PIGOTT, Robert C REINER JR., Theo VOS, Simon I HAY, Christopher J MURRAY a Stephen S LIM, 2023. Past SARS-COV-2 infection protection against re-infection: A systematic review and meta-analysis. The Lancet [online]. roč. 401, č. 10379, s. 833–842. Získáno z: doi:10.1016/s0140-6736(22)02465-5
- ↑ Emilia Vynnycky, Amy Trindall, Punam Mangtani, Estimates of the reproduction numbers of Spanish influenza using morbidity data, International Journal of Epidemiology, Volume 36, Issue 4, August 2007, Pages 881–889, https://doi.org/10.1093/ije/dym071
- ↑ Taubenberger, J. K., & Morens, D. M. (2006). 1918 Influenza: the Mother of All Pandemics. Emerging Infectious Diseases, 12(1), 15-22. https://doi.org/10.3201/eid1201.050979.
- ↑ SPINNEY, Laura, 2020. The long flu sufferers of the 1918-1919 pandemic. Time [online] [vid. 16. leden 2024]. Získáno z: https://time.com/5915616/long-flu-1918-pandemic
- ↑ EISENBERG, Joseph, 2020. R0: How scientists quantify the intensity of an outbreak like coronavirus and its pandemic potential: The pursuit: University of Michigan School of Public Health: Coronavirus: Pandemic. R0: How Scientists Quantify the Intensity of an Outbreak Like Coronavirus and Its Pandemic Potential | The Pursuit | University of Michigan School of Public Health | Coronavirus | Pandemic [online] [vid. 11. leden 2024]. Získáno z: https://sph.umich.edu/pursuit/2020posts/how-scientists-quantify-outbreaks.html
- ↑ Center for Disease Control and Prevention., 2023. Preliminary estimated influenza-related illnesses, medical visits, hospitalizations, and deaths in the United States – 2021-2022 influenza season. Centers for Disease Control and Prevention [online] [vid. 11. leden 2024]. Získáno z: https://www.cdc.gov/flu/about/burden/2021-2022.htm
- ↑ WHO, nedatováno. Influenza (seasonal). World Health Organization [online] [vid. 16. leden 2024]. Získáno z: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal)
- ↑ Guerra, F. M., Bolotin, S., Lim, G., Heffernan, J., Deeks, S. L., Li, Y., & Crowcroft, N. S. (2017). The basic reproduction number (R0) of measles: a systematic review. The Lancet. Infectious diseases, 17(12), e420–e428. https://doi.org/10.1016/S1473-3099(17)30307-9
- ↑ CENTERS FOR DISEASE CONTROL AND PREVENTION, 2019. Measles. Centers for Disease Control and Prevention [online] [vid. 11. leden 2024]. Získáno z: https://www.cdc.gov/globalhealth/newsroom/topics/measles/index.html
- ↑ WHO, nedatováno. Measles. World Health Organization [online] [vid. 16. leden 2024 b]. Získáno z: https://www.who.int/news-room/fact-sheets/detail/measles
- ↑ DISEASE PREVENTION AND CONTROL, SAN FRANCISCO DEPARTMENT OF PUBLIC HEALTH, 2019. Measles. Disease Prevention and Control, San Francisco Department of Public Health [online] [vid. 16. leden 2024]. Získáno z: https://www.sfcdcp.org/infectious-diseases-a-to-z/measles/
- ↑ CHAN‐YEUNG, Moira a Rui‐Heng XU, 2003. SARS: epidemiology [online]. listopad 2003. B.m.: Wiley. Dostupné z: doi:10.1046/j.1440-1843.2003.00518.x
- ↑ CHAN‐YEUNG, Moira a Rui‐Heng XU, 2003. SARS: epidemiology [online]. listopad 2003. B.m.: Wiley. Dostupné z: doi:10.1046/j.1440-1843.2003.00518.x
- ↑ CENTERS FOR DISEASE CONTROL AND PREVENTION, 2005. SARS. Centers for Disease Control and Prevention [online] [vid. 16. leden 2024]. Získáno z: https://www.cdc.gov/sars/about/faq.html
- ↑ WU, Li-Ping, Nai-Chang WANG, Yi-Hua CHANG, Xiang-Yi TIAN, Dan-Yu NA, Li-Yuan ZHANG, Lei ZHENG, Tao LAN, Lin-Fa WANG a Guo-Dong LIANG, 2007. Duration of antibody responses after severe acute respiratory syndrome. Emerging Infectious Diseases [online]. roč. 13, č. 10, s. 1562–1564. Získáno z: doi:10.3201/eid1310.070576
- ↑ Muzembo, B. A., Kitahara, K., Mitra, D., Ntontolo, N. P., Ngatu, N. R., Ohno, A., Khatiwada, J., Dutta, S., & Miyoshi, S. I. (2024). The basic reproduction number (R0) of ebola virus disease: A systematic review and meta-analysis. Travel medicine and infectious disease, 102685. Advance online publication. https://doi.org/10.1016/j.tmaid.2023.102685
- ↑ Patel PR, Shah Su. Ebola Virus. [Updated 2023 Jul 17]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK560579/
- ↑ WHO, nedatováno. Ebola virus disease. World Health Organization [online] [vid. 16. leden 2024 a]. Získáno z: https://www.who.int/news-room/fact-sheets/detail/ebola-virus-disease
- ↑ RIMOIN, Anne W, Kai LU, Matthew S BRAMBLE, Imke STEFFEN, Reena H DOSHI, Nicole A HOFF, Patrick MUKADI, Bradly P NICHOLSON, Vivian H ALFONSO, Gerrard OLINGER, Cyrus SINAI, Lauren K YAMAMOTO, Christina M RAMIREZ, Emile OKITOLONDA WEMAKOY, Benoit KEBELA ILLUNGA, James PETTITT, James LOGUE, Richard S BENNETT, Peter JAHRLING, David L HEYMANN, Peter PIOT, Jean Jacques MUYEMBE-TAMFUM, Lisa E HENSLEY a Graham SIMMONS, 2017. Ebola virus neutralizing antibodies detectable in survivors of theyambuku, Zaire outbreak 40 years after infection. The Journal of Infectious Diseases [online]. roč. 217, č. 2, s. 223–231. Získáno z: doi:10.1093/infdis/jix584